library(tidyverse)
library(lubridate)
library(sf)
library(here)
library(readr)
library(tidyr)
library(tigris)
library(rmapshaper)
library(mapview)About
Bigfoot Field Researchers Organization (BFRO) organizes and reports observations and directs expeditions to places where the observations have occurred. The overall mission of the BFRO is multifaceted, but the organization essentially seeks to resolve the mystery surrounding the bigfoot phenomenon, that is, to derive conclusive documentation of the species’ existence. This goal is pursued through the proactive collection of empirical data and physical evidence from the field and by means of activities designed to promote an awareness and understanding of the nature and origin of the evidence. See more information about the organization here.
Load packages
Load in Bigfoot sightings data
bigfoot_raw <- readr::read_csv(here("posts", "2024-03-15-bigfoot-exploration", "bfro_reports_geocoded.csv")) |>
dplyr::select(county, state, latitude, longitude, date) |>
tidyr::drop_na() |>
mutate(year = year(date)) |>
filter(!state %in% c("Alaska", "Hawaii"),
!county %in% c("Idaho County", "Cowlitz County", "Flathead County")) # drop these random counties that are spatially incorrect
head(bigfoot_raw)# A tibble: 6 × 6
county state latitude longitude date year
<chr> <chr> <dbl> <dbl> <date> <dbl>
1 Wyoming County West Virginia 37.6 -81.3 2005-12-03 2005
2 Windsor County Vermont 43.5 -72.7 2005-10-08 2005
3 Wythe County Virginia 37.2 -81.1 1984-04-08 1984
4 Wood County Texas 32.8 -95.5 1996-12-22 1996
5 Washington County Rhode Island 41.4 -71.5 1974-09-20 1974
6 Washita County Oklahoma 35.3 -99.2 1973-09-28 1973Make lat and long spatial features and summarize
# Define the coordinate reference system (CRS)
crs <- st_crs(4326)
# Convert to an sf object
bigfoot_sf <- bigfoot_raw |>
st_as_sf(coords = c("longitude", "latitude"),
crs = crs)
# Summarize the sightings by county
bigfoot_summary_df <- bigfoot_sf |>
group_by(county) |>
summarise(number_of_sightings = n()) |>
ungroup() Add conus sf for plotting
conus_sf <- tigris::states(cb = TRUE) |>
st_transform(crs) |>
mutate(group = case_when(
STUSPS %in% c(state.abb[!state.abb %in% c('AK', 'HI')], 'DC') ~ 'CONUS',
STUSPS %in% c('GU', 'MP') ~ 'GU_MP',
STUSPS %in% c('PR', 'VI') ~ 'PR_VI',
TRUE ~ STUSPS
)) |>
filter(group %in% c('CONUS')) |>
rmapshaper::ms_simplify(keep = 0.2)
counties_conus_sf <-tigris::counties() |>
st_transform(crs = crs) |>
left_join(conus_sf |>
st_drop_geometry() |>
dplyr::select(STUSPS, STATE_NAME = NAME, STATEFP, group),
by = 'STATEFP') |>
rmapshaper::ms_simplify(keep = 0.2) |>
st_intersection(st_union(conus_sf)) |>
rename(county = NAMELSAD)Quick plot of data points
counties_outline_col = "grey80"
conus_outline_col = 'grey50'
ggplot() +
geom_sf(data = conus_sf,
fill = NA,
color = conus_outline_col ,
linewidth = 0.2,
linetype = "solid") +
geom_sf(data = counties_conus_sf,
fill = NA,
color = counties_outline_col ,
linewidth = 0.05,
linetype = "solid") +
geom_sf(data = bigfoot_sf,
color = "darkgreen",
size = 2,
alpha = 0.5) +
theme_void() 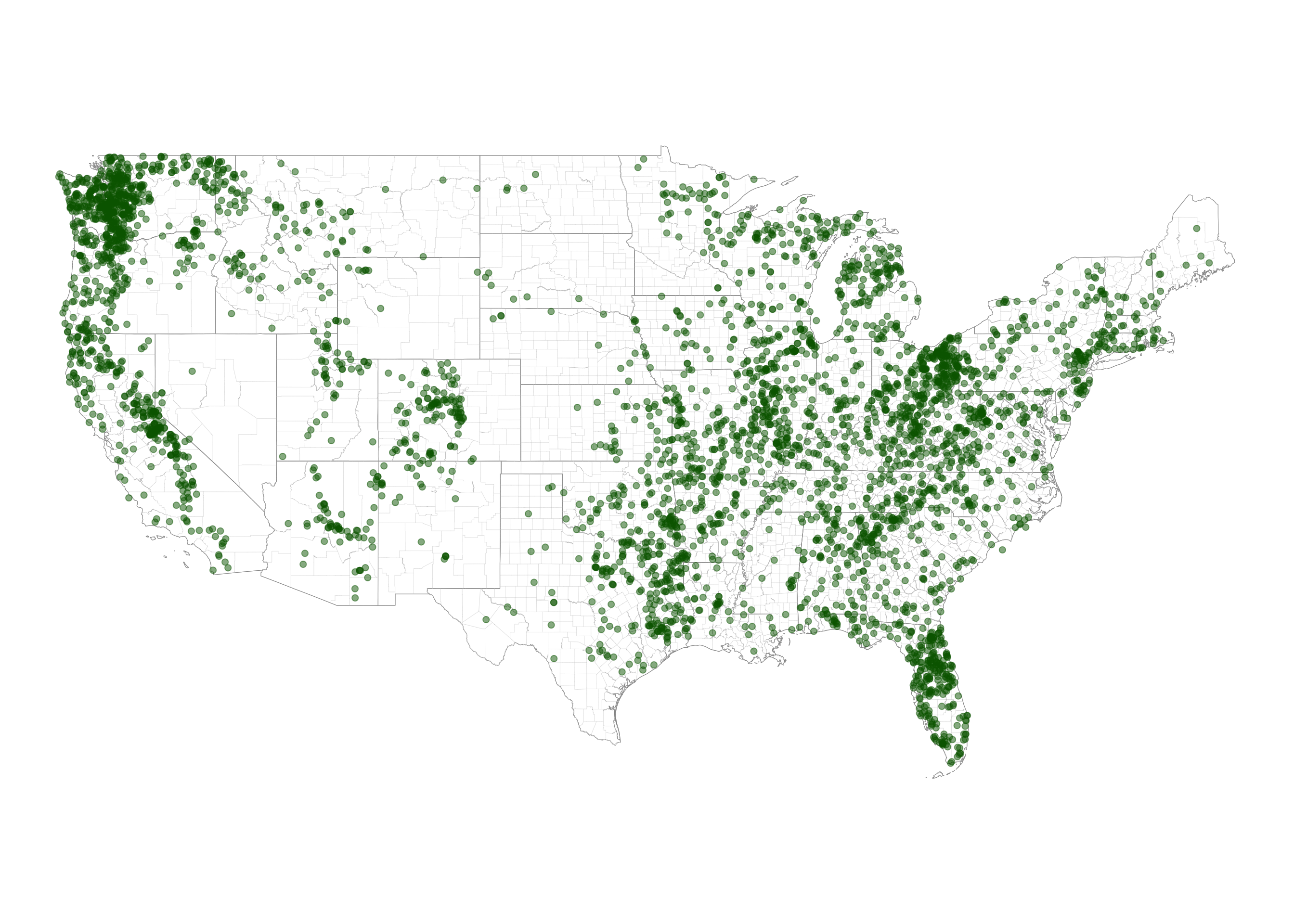
Interactive view of Bigfoot sightings
mapview::mapview(bigfoot_sf, alpha = 0.5, col.regions ="darkgreen",
layer.name = "Bigfoot Sightings from 1869-2023",
map.types = c("CartoDB.DarkMatter", "Esri.WorldImagery"))